Tyler Ford
06/10/2019
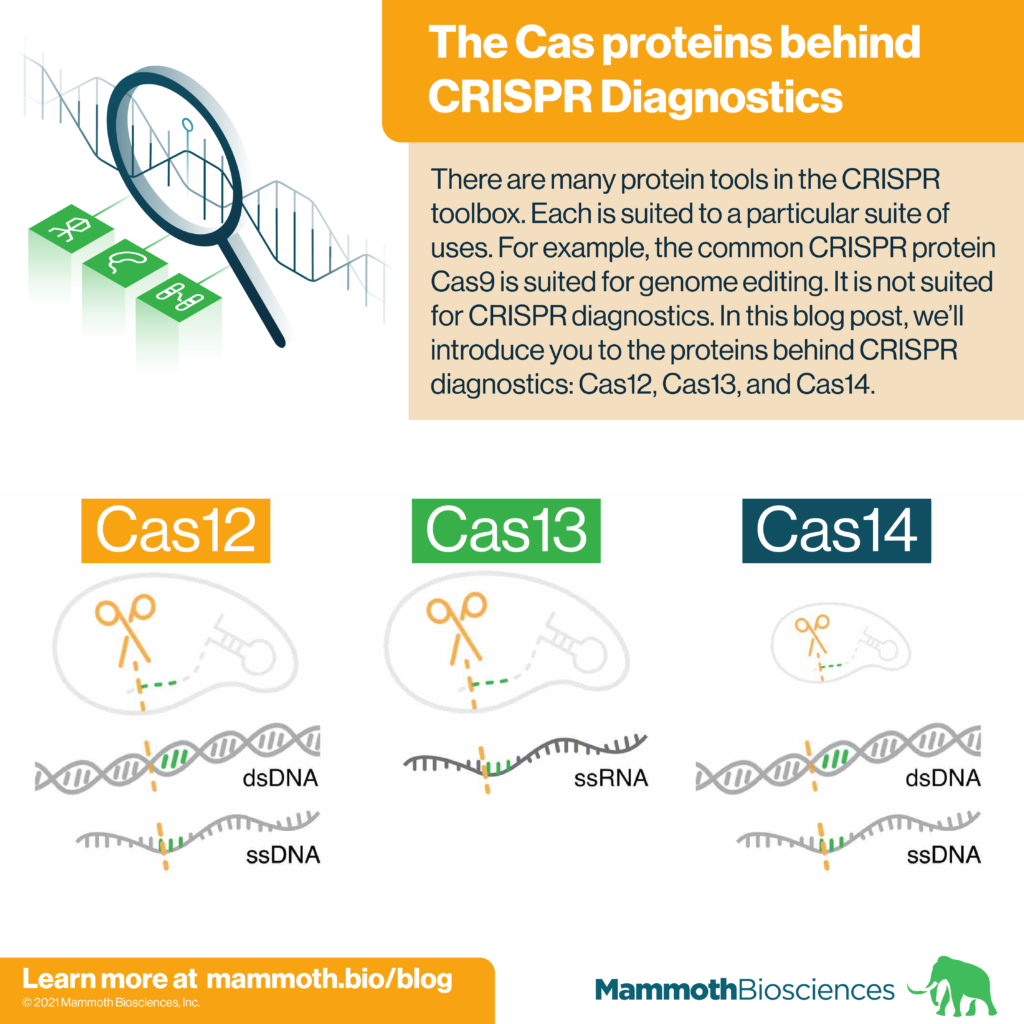
There are many protein tools in the CRISPR toolbox. Each is suited to a particular suite of uses. For example, the common CRISPR protein Cas9 is often used for genome editing. It is not suited for CRISPR diagnostics. In this blog post, we’ll introduce you to the proteins behind CRISPR diagnostics: Cas12, Cas13, and Cas14.
CRISPR diagnostics make use of non-specific cutting
CRISPR diagnostics have two key components:
- Protein-guide molecule complexes. These first cut specific nucleic acid sequences that the user wants to detect. After cutting a user-specified sequence, these complexes non-specifically cut other nucleic acids.
- Modified nucleic acids (reporters). These produce a visual signal when cut. They are only cut if the user-specified nucleic acids are cut first. These modified nucleic acids make it easy to observe when the user-specified nucleic acids have been detected (cut).
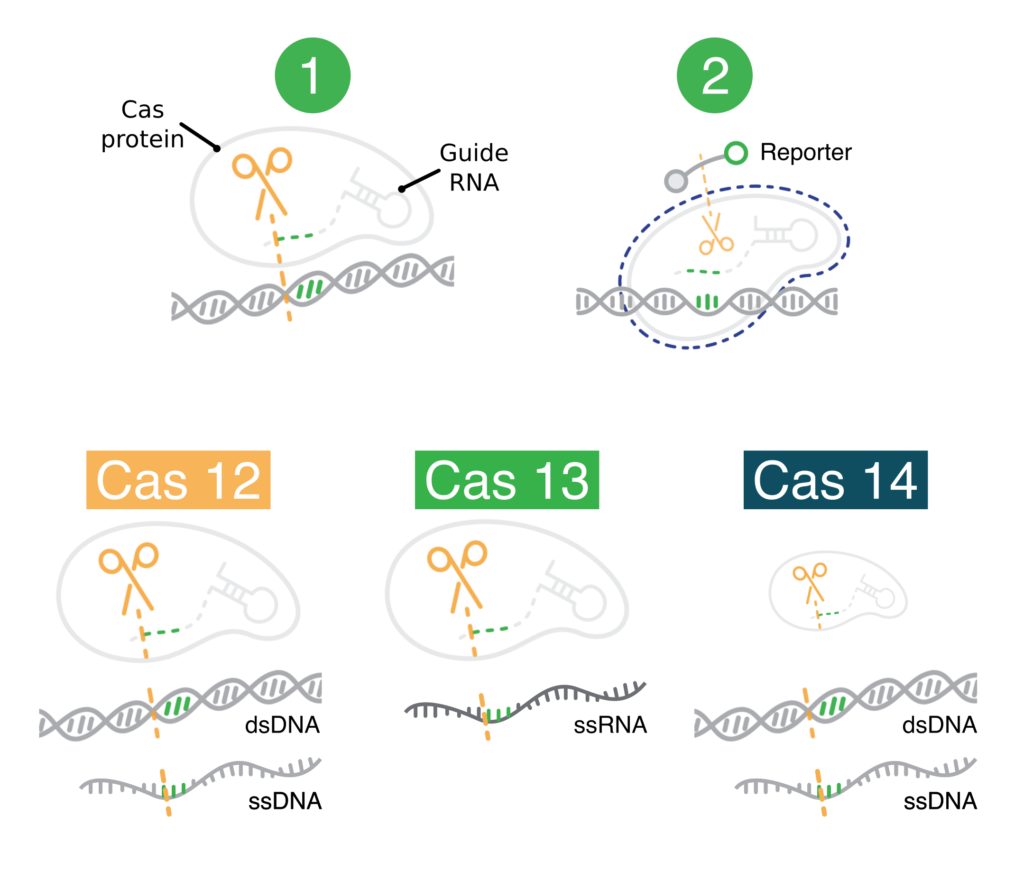
Bottom: The Cas12, Cas13, and Cas14 protein-guide molecule complexes. These are capable of cutting the indicated types of user-specified nucleic acid sequences. In a CRISPR diagnostic, these would go on to cut modified nucleic acids nonspecifically.
In diagnostics, it’s critical that non-specific cutting comes after specific cutting. Nonspecific cutting (sometimes called “collateral,” “trans,” or “indiscriminate”) results in cleavage of the modified nucleic acids. The visual signal produced by the modified nucleic acids then shows that the user-specified sequence has been detected. If non-specific cutting came first, CRISPR diagnostics would always produce a visual signal. They would be useless.
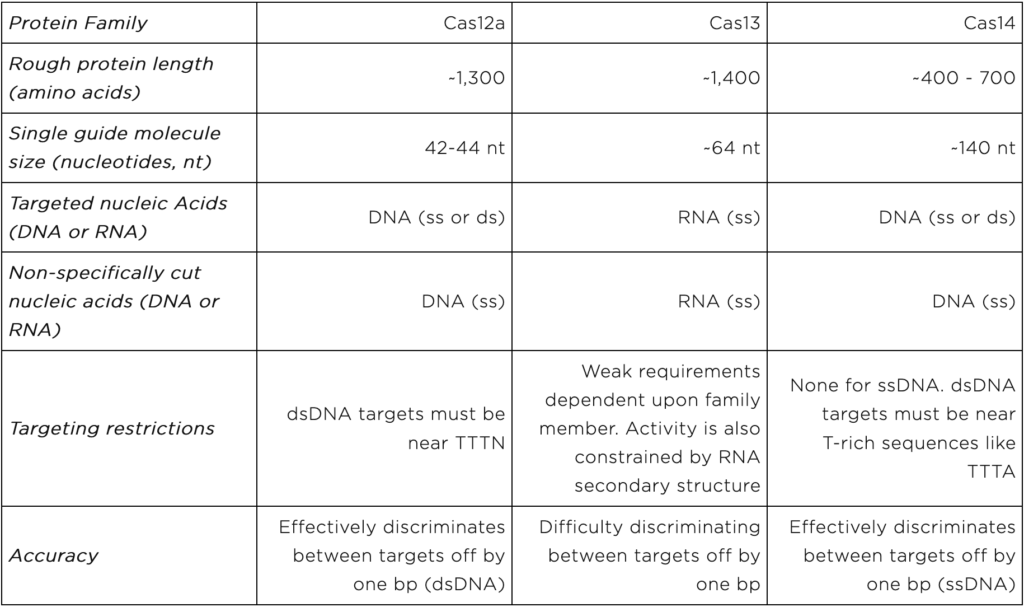
Cas12, Cas13, and Cas14 are families of proteins used in CRISPR diagnostics. They form the protein portion of the “protein-guide molecule complexes” described above. Individual family members come from specific species of bacteria and archaea. Yet, all members of a given family share certain characteristics. Characteristics important for CRISPR diagnostics are displayed in table 1 and discussed below.
Cas12
The Cas12 proteins directly bind to and cut user-specified DNA sequences. They can cut either single or double stranded DNA. Once a Cas12 protein cuts its DNA target, it begins to shred single stranded DNA non-specifically. Thus Cas12-based diagnostics can only directly detect DNA. They must be combined with proteins that convert RNA into DNA to detect RNA.
Cas12 proteins are on the larger side of the CRISPR diagnostic proteins. They come in at ~1,300 amino acids long.
Users specify Cas12 DNA targets using 42-44 nt RNA molecules.
Cas12 dsDNA targets are restricted in that they must be found near short stretches of DNA known as protospacer adjacent motifs (PAMs). For some Cas12 proteins, the PAM sequence is TTTN. Importantly, Cas12-based diagnostics cannot detect DNA sequences without PAMs.
Cas12 proteins can readily distinguish very similar dsDNA sequences. This feature is lost when the target sequence is ssDNA.
Cas13
The Cas13 proteins directly bind and cut user-specified RNA sequences. After cutting a target, Cas13 proteins non-specifically cut other RNA molecules. Thus, they can directly detect RNA, but not DNA. To detect DNA, Cas13-based diagnostics must be combined with proteins that convert DNA into RNA.
Like Cas12, Cas13 proteins are on the larger end of the CRISPR diagnostic proteins at ~1,400 amino acids long.
Like Cas12, the Cas13 RNA guide molecule is relatively short at ~64 nt.
Cas13 proteins do not have strong targeting restrictions. Yet, their RNA targets can adopt structures that are difficult to cut. These structural constraints limit the targets detectable by Cas13-based diagnostics.
Cas13 will cut RNA sequences that are 1 nt off from the user-specified sequence. Thus, researchers must carefully test Cas13-based diagnostics when using them to distinguish between very similar sequences.
Cas14
The Cas14 proteins bind to and cut user-specified, single-stranded or double stranded DNA. To detect RNA, Cas14-based diagnostics must be combined with proteins that convert RNA into DNA.
The Cas14 proteins are on the smaller end at 400 – 700 amino acids. However their guide RNAs are on the longer side at ~140 nt.
Cas14 proteins have no targeting restrictions when cutting ssDNA. They are highly versatile. For dsDNA targeting, Cas14 proteins require T-rich PAM sequences like TTTA .
Cas14 proteins can readily distinguish between very similar ssDNA sequences.
Combined applications and the future of CRISPR diagnostic proteins
As you can see, each of these protein families has its own pros and cons. Importantly, researchers have creative ways to combine proteins from multiple families in single detectors. They can use combinations of proteins to detect multiple targets at once.
Scientists discover and alter new CRISPR systems all the time. They’re sure to find and create more CRISPR diagnostic tools. These tools will have applications in healthcare, agriculture, food security, infectious diseases, and beyond. At Mammoth we’re dedicated to expanding the CRISPR toolkit and broadening the impact of the CRISPR revolution. Stay tuned!
Table 1: Cas12, Cas13, and Cas14 properties important for CRISPR diagnostics
Learn why “Point-of-Need” diagnostics are essential for fighting infectious disease!
