Tyler Ford
03/09/2021
Cas proteins power the CRISPR genome editing revolution. Indeed, it is Cas proteins, along with guides, that target and cut specific DNA sequences during the genome editing process.
Two popular Cas proteins used for genome editing are SpCas9 and its smaller cousin, SaCas9. Although these two proteins enable many genome editing efforts, they do have drawbacks. Here, we focus on Cas9 immunity. That is, humans may mount immune responses to Cas9 proteins. This is problematic for two main reasons:
- Immune responses to Cas9 may limit the effectiveness of genome editing. For example, the immune system may kill off cells that express Cas9.
- Immune responses to Cas9 may cause serious side effects in patients administered Cas9 as part of a genome editing therapy.
Below, we discuss some of the reasons and evidence for Cas9 immunity. We also discuss solutions to Cas9 immunity. In particular, at Mammoth, we’re attempting to mitigate the Cas9 immunity problem by developing alternative Cas proteins for genome editing.
Immune responses to Cas9
As their names indicate, SpCas9 and SaCas9 come from the bacteria Streptococcus pyogenes and Staphylococcus aureus, respectively. Unfortunately, both of these bacteria are opportunistic pathogens in humans. S. pyogenes can cause pharyngitis, scarlet fever, and rheumatic fever (Efstratiou 2017). S. aureus can cause a wide range of healthcare-associated and skin infections (e.g. MRSA, boils, cellulitis) (Lowy 1999, Tong et al 2015).
Many of us come into contact with S. pyogenes and S. aureus through the course of normal life (Efstratiou 2017, Tong et al 2015). As a result, Charlesworth et al 2019 hypothesized that some fraction of the human population might have pre-existing immunity to SpCas9 and SaCas9. To test this hypothesis, they searched for antibodies against SpCas9 and SaCas9 in donated human blood. They also checked to see if the donated blood contained immune cells (e.g., T-cells) that specifically recognize SpCas9 and SaCas9. They found that more than 50% of donors had antibodies to SpCas9 or SaCas9. In addition, more than 60% of donors had T-cells directed against SpCas9 or SaCas9.
This finding and others like it (Federosi et al 2019, Wagner et al 2019, Simhadri et al 2018) do not prove that these immune responses will cause harmful side effects or limit the impact of genome editing in humans. However they do raise concerns meriting additional research.
Indeed, Li et al 2020 recently found that SaCas9 can trigger the immune system in detrimental ways in mice. In their work, they assessed the impact of SaCas9 exposure on genome editing. Because they worked with lab mice that were never exposed to Staphylococcus aureus before, they first inoculated their mice with SaCas9 or a control protein. They found that these mice generated immune responses to SaCas9 including inflammation and immune memory.
These researchers then specifically checked if the observed immune responses could impact genome editing. They inoculated new mice with SaCas9 or a control protein. Later, they administered the mice viral vectors expressing SaCas9 and targeting the ldlr gene for genome editing in the liver. Then, they checked how effectively the ldlr gene was edited in liver cells. They compared liver cells from mice that were pre-inoculated with SaCas9 to those pre-inoculated with a control protein.
The mice that had been pre-inoculated with SaCas9 showed signs of liver damage and ultimately had fewer genome-edited cells. Thus, at least in mice, SaCas9 immunity appears to limit the impact of genome editing.
It is unclear whether genome editing experiments will encounter similar problems in humans. Successful CRISPR-based therapies for blood disorders to-date use cells that were edited outside the body and later inserted into patients. These pre-edited cells may not elicit strong immune responses. Therapies that either use viral vectors to facilitate Cas9 expression in the body or which deliver Cas9 protein directly to cells in the body may elicit responses more similar to those found in mice.
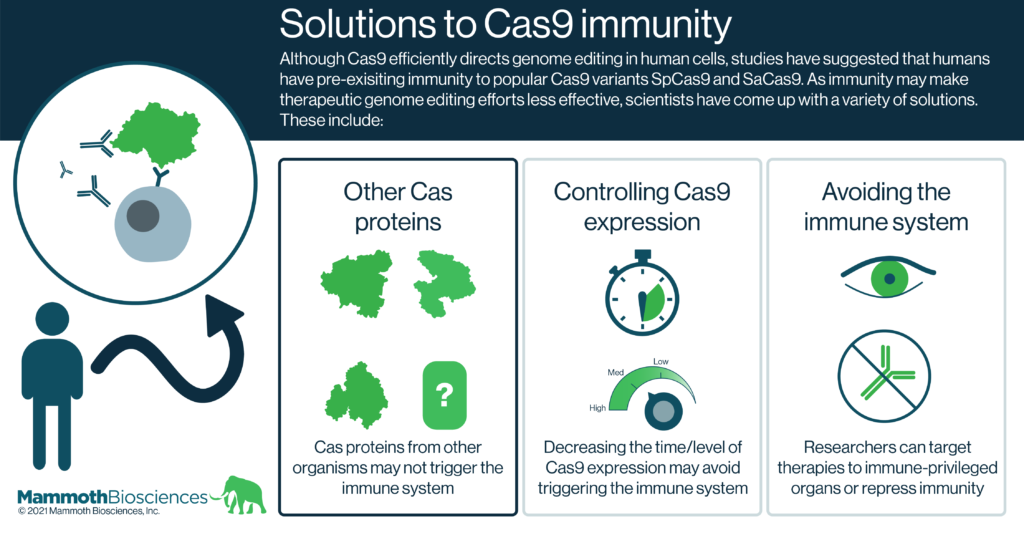
Solutions to Cas9 immunity
Forseeing the potential problem of Cas9 immunity in CRISPR therapies, researchers have come up with many solutions. Here we group these solutions into 3 broad categories:
- Natural and engineered alternatives to Cas9
At Mammoth, we are developing new Cas proteins for genome editing. These include Cas14 and CasPhi. Many of these nucleases come from organisms that are not human pathogens. Thus our immune systems are unlikely to have encountered these proteins in the past and may not mount strong responses to them upon first administration.
Other researchers are working to determine which portions of Cas9 trigger the immune system. They are using protein engineering techniques to alter or remove these portions of Cas9. Proof of concept studies show that these engineered Cas9 variants are less immunogenic as a result (Federosi et al 2019).
- Controlling Cas9 expression
If cells express Cas9 only when and where it is needed or at levels that direct genome editing but fail to elicit immune responses, worries about Cas9 immunogenicity may be mitigated. Indeed, there are many ways to control when and where Cas9 is expressed. These include:
Inducible promoters – Promoters are DNA sequences that control when, where, and to what level a gene is expressed. Some promoters can be “turned on” by specific small molecules and may even be turned up or down with these small molecules.
Specially designed viral vectors – Researchers can use viruses to deliver genes to cells. Some viruses can be designed to deliver genes to specific types of cells. Some viral vectors will degrade over time. Using the right viral vectors, researchers can limit Cas9 expression to specific cells and periods of time.
Ribonucleoproteins (RNPs) and lipid nanoparticles (LNPs) – Rather than delivering Cas9 DNA to cells that later decode the DNA to produce Cas9 protein, researchers can directly deliver Cas9 and its associated guide RNAs to cells in the form of RNPs or LNPs. They stick around in cells for a limited period of time before being degraded. They can also be delivered to specific parts of the body.
- Immune suppression and delivery to immune-privileged organs
Researchers and physicians have been dealing with immune responses to gene therapies for many years. Indeed, one of the most popular gene therapy delivery vehicles, adenovirus associated vectors (AAVs), commonly elicits adaptive immune responses. To get around this issue, gene therapy researchers are looking for ways to suppress the adaptive immune system and prevent responses to AAVs. Similar things could be done when scientists deliver Cas9 to patients.
Even without immune suppression, some parts of the body are immune-privileged. That is, the immune system does not mount strong responses in these parts of the body. The human eye is one such immune-privileged organ and therapeutic genome editing efforts are already targeting cells in the eye (e.g., this therapy targeting Leber’s congenital amaurosis 10). Such therapies are unlikely to have issues with Cas9 immune responses.
While all of the above are workable solutions to the problem of Cas9 immunity, at Mammoth, we believe the simplest solution is to use alternatives to Cas9 for therapeutic purposes. Thus, we are proud to be developing a unique and diverse set of Cas proteins for genome editing!
