Tyler Ford
10/19/2020
Genome editing is the process researchers use to make targeted changes to an organism’s DNA (its genome). Scientists have used a variety of technologies for genome editing (see the history of genome editing here). However, since ~2012, CRISPR has made the genome editing processes more modular and adaptable. CRISPR associated or “Cas” proteins drive this process. They are relatively easy to target to specific DNA sequences. They also work in many organisms.
Yet, the main Cas protein currently used for CRISPR genome editing, SpCas9, has limitations. In this post, we cover SpCas9’s limitations and how newly discovered Cas protein families, Cas14 and CasΦ, potentially overcome these limitations. We hope Cas14 and CasΦ will enable more efficient genome editing in diverse organisms and tissues.
The limitations of SpCas9
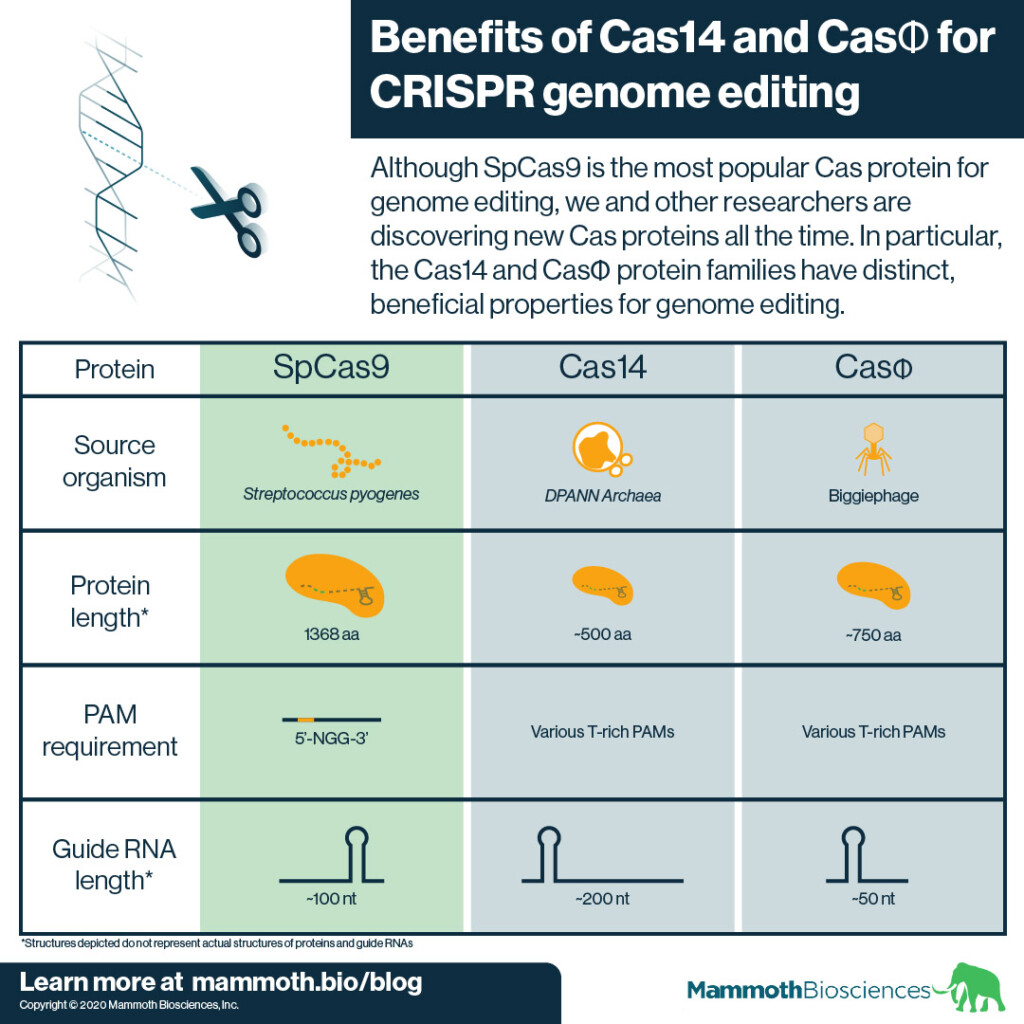
SpCas9 comes from a bacterium called Streptococcus pyogenes (hence “Sp”). S. pyogenes is a human pathogen. There is some evidence that using SpCas9 for genome editing in humans may lead to dangerous immune reactions (Ferdosi et al 2019) although other reports have questioned the importance of this finding.
In addition SpCas9 is quite large. It is 1368 amino acids (aa) long and can be difficult to fit into standard delivery vehicles (learn about CRISPR delivery here). Thus it can be hard to get SpCas9 into target cells and tissues.
Finally, SpCas9 requires the presence of a specific DNA sequence known as a “PAM” to target an adjacent sequence for genome editing. SpCas9’s PAM is 5’-NGG-3’. The need for this sequence restricts the number of sites SpCas9 can edit. This limits SpCas9’s usefulness.
Metagenomics researchers discover Cas14 and CasΦ
Given the restrictions of SpCas9, researchers have been using metagenomics to discover new Cas proteins for genome editing and other applications. You can read more about the metagenomics discovery process here. Briefly, this process makes use of DNA sequence databases sampled from many environments. Researchers sift through these sequences to find signatures of CRISPR systems. Once found, they isolate these systems and determine if they are functional. Some functional systems can later guide genome editing.
Using metagenomics techniques, our Co-Founder and CSO, Lucas Harrington, and his colleagues found and characterized Cas14. One of the members of the Cas14 protein family came from an archaeon sampled by the Banfield Lab from ground water near Rifle Colorado. Fun fact – researchers study microorganisms sampled from Rifle because it is near historical uranium and vanadium mines. By studying these microorganisms, researchers hope to understand how residual contamination from the mines affects the environment.
Similarly, Co-Founder and Nobel Laureate Professor Jennifer Doudna and colleagues recently discovered and characterized CasΦ. This Cas protein family comes from viruses that infect bacteria, “phages.” In particular, CasΦ came from a group of phages with large genomes known as “Biggiephages”. These phages have been found in metagenomic samples from a variety of locations. It’s believed that phages use CRISPR systems to disrupt certain bacterial processes. They may also prevent other phages from infecting the same bacterial host (Al-Shayeb et al 2020).
Benefits of Cas14 and CasΦ for genome editing
As outlined in the graphic above, Cas14 and CasΦ both have beneficial properties for genome editing. These properties may make them more useful than SpCas9.
To begin with, neither Cas14 nor CasΦ comes from a human pathogen. Thus, they may be less likely than SpCas9 to trigger immune responses when used in humans.
In addition, both Cas14 and CasΦ are quite small. Members of the Cas14 family come in around 500 aa. Members of the CasΦ family are slightly larger at around 750 aa. This means proteins from both families should be easier to deliver to target tissues than SpCas9.
Finally, both Cas14 and CasΦ have less restrictive PAM requirements than SpCas9. They use T-rich sequences as PAMs. For example, one of CasΦ’s PAMs is 5’-TBN-3’ (where B is G,T, or C). These loose PAM requirements open more DNA sequences to genome editing by Cas14 and CasΦ thus making them more useful than SpCas9.
Using Cas14 and CasΦ
You can learn about some of the many applications of genome editing in healthcare and crops in the linked blog posts. We hope these proteins will make it easier to achieve a variety of biotechnological goals and we’re looking for partners! If you’d like to partner with us to develop Cas14 or CasΦ for a particular application, please reach out via the form found on our Partner Page.
