Tyler Ford
04/13/2020
Only a small fraction of the world’s microorganisms have been cultured. Yet, we do have large databases of DNA sequences from a wide array of organisms. In a previous blog post, we discussed how scientists use such databases to discover new CRISPR systems. This work comprises a small part of the field of “metagenomics.”
In metagenomics, researchers use computational tools to search through DNA databases and find sequences encoding proteins, groups of proteins, or even new organisms. Using metagenomics, we can find many new kinds of tools with biotechnological applications.
At Mammoth, we’re expanding our metagenomics expertise and capabilities through our protein discovery platform. In this post, we’ll give you a better feel for why metagenomics is so useful. Below we highlight some of the exciting discoveries that have come from metagenomics.
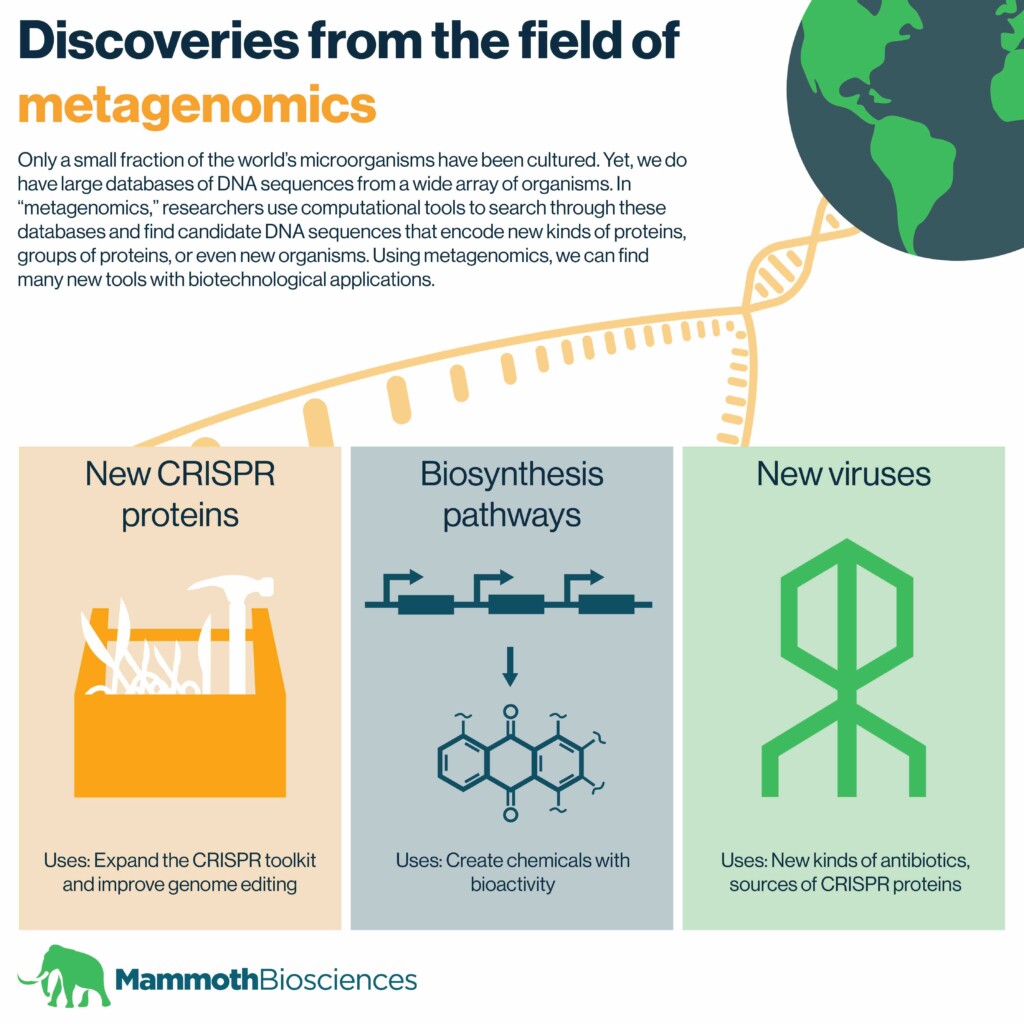
A slew of CRISPR proteins
We are using metagenomics techniques to discover new CRISPR systems. In doing so, we will gain access to powerful new CRISPR-associated (Cas) proteins.
Researchers have used metagenomics to discover many Cas proteins. Some of these include:
- Cas14 – Cas14 is one of the key Cas proteins behind our CRISPR diagnostics. It is also smaller than many other Cas proteins. The extremely small size of Cas14 family members may be one of several advantages for their use in genome editing. This is especially true for in vivo genome editing where size can be particularly limiting.
- CasY – Like Cas14, CasY has the collateral cutting activity required for CRISPR diagnostics. It is also compact and has very simple requirements for targeting DNA. This may make CasY a more versatile genome editor than many other Cas proteins.
- Cas12j – Unlike the above Cas proteins, Cas12j comes from the viruses that infect bacteria (phage). It has the collateral cutting activity required for CRISPR diagnostics and is more compact than many of the Cas proteins commonly used for genome editing.
- GeoCas9 – GeoCas9 was found in the genome of a thermophilic (heat loving) bacteria native to hot springs. It is called Geobacillus stearothermophilus (hence “GeoCas9”). GeoCas9 functions best at temperatures from 50 – 70 °C. This may make GeoCas9 particularly useful for genome editing in thermophiles. It may also be useful for applications that proceed at high temperatures.
Many other Cas proteins have been discovered using metagenomics. We’re excited to direct our expertise toward finding even more!
Novel biosynthetic gene clusters
There is great interest in looking to nature for chemicals with useful properties. We can make many useful chemicals in labs. Yet, nature has been tinkering with chemistry since the dawn of life. As such, many organisms produce arrays of chemicals with useful functions. Some of these are “bio-active” and affect the biology of other organisms .
Indeed, nature produces many antimicrobials – compounds that kill microorganisms. Some of these can kill microorganisms that cause infectious disease. As antibiotic resistance becomes more prevalent, it will become ever more important to look to nature for new antimicrobials. By searching through natural systems, we should be able to find new chemicals that enable us to fight off infectious disease.
Toward this end, researchers recently used metagenomics techniques to identify “biosynthetic gene clusters”. These clusters consist of genes encoding proteins known to synthesize chemicals. In this particular study, researchers looked for biosynthetic gene clusters encoding Type II polyketide synthases. These researchers searched the genomes of organisms from across the globe, found in animals, and even found under the sea.
After finding many such gene clusters, the researchers expressed the clusters in tractable lab organisms. As a result, the lab organisms produced a variety of new chemicals. Some of these chemicals even had potent antimicrobial activities.
Hopefully many more similar studies will identify more bio-active compounds!
Lots of new viruses – Many of them huge!
We know very little about the viruses of the world. This is unfortunate because, in their constant struggle to infect new hosts, viruses continuously create new tools. These tools enable viruses to manipulate host biology in ways that may be useful to us.
For instance, many viruses (phage) infect bacteria. Indeed, there are efforts to harness phage attack bacteria and treat bacterial infections. Some of these have already been successful. If we can find and learn enough about phage to understand what makes them potent bacterial killers, we can more effectively use them to fight disease. They’ll become new tools in our arsenal against antibiotic resistance.
Thankfully, researchers have made great strides in using metagenomics to discover new viruses. In 2016, Paez-Espino et al created a pipeline for virus discovery. They found many new viruses that infect microorganisms. One of the lead authors on this work, David Paez-Espino, is now a member of the Mammoth Team as our Associate Director of Discovery Informatics.
More recently, researchers from the Innovative Genomics Institute used their own metagenomics pipeline to identify “huge phages.” These are phages with large genomes (for phages). Intriguingly, many huge phages have their own CRISPR systems. They could be yet another source of tools for the CRISPR toolkit!
More metagenomics to come
These are just some of the amazing discoveries coming out of metagenomics. As we gain access to genetic sequences from more organisms and improve our analysis pipelines, we’re sure to make many more exciting discoveries!
If you’d like to partner with us in our metagenomics efforts, please reach out here.
