Tyler Ford
09/17/2019
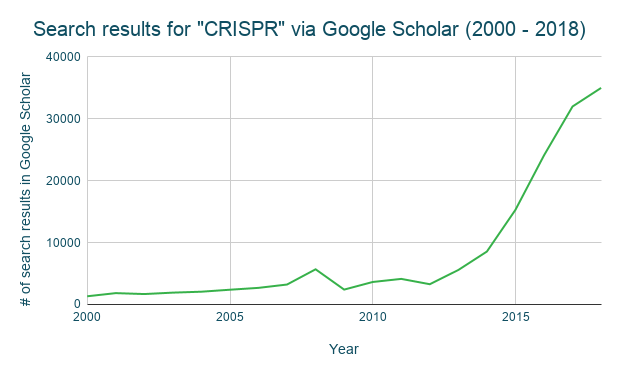
CRISPR research has experienced rapid growth since CRISPR genome editing hit the scene back in 2012. If you take a look at the number of scholarly articles containing the word “CRISPR” from 2000 – 2018, the increase after 2012 is astounding:
The discovery of many different CRISPR systems from many different microbes has come along with this explosion of research. These systems evolved to help microbes fight off a wide array of viruses. Thus, CRISPR systems are quite diverse but generally have two main components: guides and effectors. Guides direct effectors to specific nucleic acid (DNA or RNA) sequences. Effectors carry out functions once they reach such sequences. Often the function is cutting apart the sequence.
The original CRISPR genome editing tools make use of a particular CRISPR system from the bacterium Streptococcus pyogenes. This system contains a CRISPR associated (Cas) protein called Cas9. It cuts apart targeted DNA as directed by a guide RNA molecule. Cas9 guide RNAs are easy for researchers to design. Cas9 itself is also quite good at cutting targeted DNA. Thus, even today, Cas9 is widely used by many researchers. It is also a key component of some of the biggest CRISPR patent disputes.
Despite Cas9’s dominance, there are many many other CRISPR systems. These make use of an astounding array of Cas proteins. At Mammoth, we’re dedicated to expanding the CRISPR toolbox beyond Cas9. The reason? Other CRISPR systems have diverse properties that make them useful for different applications. We discuss some of the diverse properties of the expanded CRISPR toolbox below.
Diverse properties of the expanded CRISPR toolbox
Guide and effector complexity – Guides and effectors from different CRISPR systems come in varying degrees of complexity. Some guides and effectors have multiple components. Some guides and effectors require processing before reaching full functionality. All else being equal (it rarely is) simpler is better.
Size- Guides and effectors come in varying sizes in terms of number of nucleotides and number of amino acids respectively. The bigger the effector-guide combo, the more difficult it is to package and deliver to cells. Smaller is usually better.
Cutting – Some CRISPR systems make blunt cuts in DNA. Some CRISPR systems make staggered cuts in DNA. Other CRISPR systems nick DNA. Still others shred DNA apart.
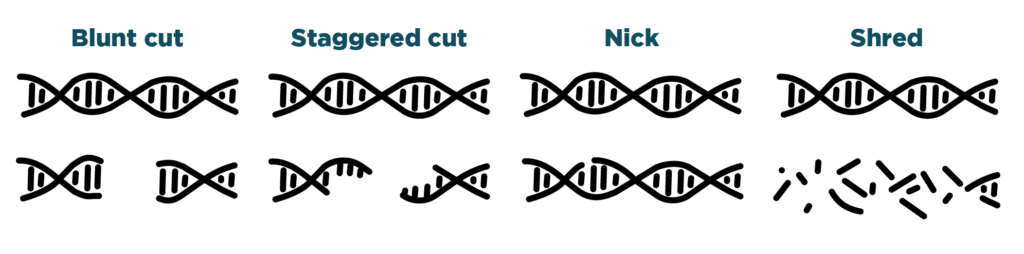
Researchers want different kinds of cuts depending upon on how they’re using their CRISPR systems. For example, if you want to break a single gene in a cell, a blunt cut might be the best choice. If you want to make a precise change to a gene, staggered cuts might be better.
It all depends on the application. Thus having access to many different systems in the expanded CRISPR toolbox is beneficial.
Targeting – Some CRISPR systems cut DNA. Some cut RNA. Others cut both DNA and RNA. Which of these a researcher wants depends on the application. For example, if you’re trying to alter gene expression without changing a gene’s underlying DNA sequence, you’ll want a CRISPR system that effectively cuts RNA but not DNA.
Other factors also restrict targeting. For example, many CRISPR systems require specific, short nucleotide sequences next to their targets. These “protospacer adjacent motifs (PAMs)” vary from system to system. Often, if a PAM is not present near a target, the effector will not cut. Having access to many different CRISPR systems means a researcher is less limited by PAM requirements.
Finally, some CRISPR systems cut nucleic acids indiscriminately after cutting a guide-specified target. This can be useful in certain applications (e.g. diagnostics).
Accuracy – Sometimes CRISPR systems act on the wrong nucleic acid sequences. Different systems have different propensities for such “off-target” effects. However, increased accuracy can come with other trade-offs. A highly accurate CRISPR system may be very large, have lower on-target editing, or might have very restricted targeting. Having access to the wide array of CRISPR systems in the expanded CRISPR toolbox allows researchers to weigh the importance of such tradeoffs. Then they can choose the best systems for their specific applications.
The CRISPR toolbox at Mammoth
At Mammoth, we’re proud to be working with and developing a large CRISPR toolbox. In so doing, we hope to ensure CRISPR’s usefulness for a wide variety of applications. If you’d like to partner with us for CRISPR tool development, let us know!
