Tyler Ford
01/28/2020
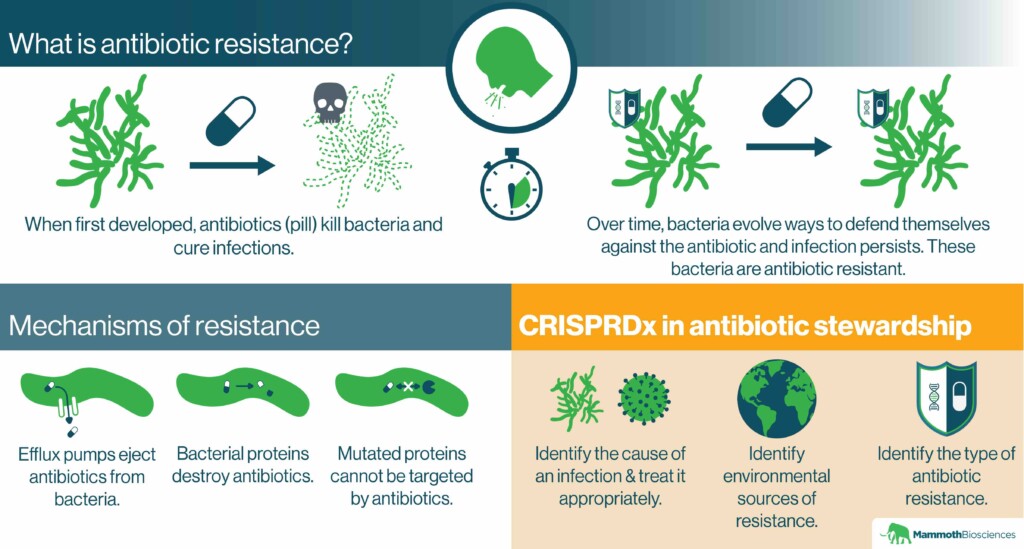
Many of the microorganisms that cause disease cannot be killed with common antibiotics. They are “antibiotic resistant.” This makes it difficult for physicians to ward off what were once easy-to-treat infections. In the U.S., antibiotic resistant infections cause 35,000 deaths per year. Unfortunately, the number of illnesses and deaths due to antibiotic resistant infections is expected to increase dramatically by 2050.
Part of the solution to this problem is to develop new antibiotics. However, this is an expensive process. Even with mounting urgency, there aren’t many antibiotics in development. In addition, developing, distributing, and selling new antibiotics in regions with poor healthcare infrastructure may be difficult.
As a result, antibiotic stewardship is essential. That is, we should find ways to use current antibiotics sparingly. We should only use them to treat infections that they’ll be effective against. Read on to learn how CRISPR diagnostics will make us better stewards of our antibiotic arsenal.
How does antibiotic resistance arise?
Microorganisms evolve antibiotic resistance naturally. Yet, this evolution is accelerated through the overuse of antibiotics. Once evolved, microorganisms can rapidly share their defenses with one another.
One example of overuse is prescribing antibiotics for infections that they cannot fight. For instance, antibiotics might be prescribed for viral infections despite the fact that antibiotics have no effect on viruses. The CDC estimates that similar unnecessary uses of antibiotics make up 30% of hospital prescriptions.
Physicians sometimes prescribe antibiotics unnecessarily because standard diagnostic tests are too slow. Rather than wait for a diagnostic test to identify the cause of an infection, physicians prescribe antibiotics “just in case.” This can prevent patients from getting sicker in the time required for the test. While potentially effective in the short term, this exposes more microorganisms to antibiotics than necessary. It pushes resistance to evolve sooner.
Without a specific diagnosis, a physician may also prescribe an antibiotic that can kill a wide range of bacterial species. Such “broad spectrum” antibiotics are effective against many infections, but their use also prompts the evolution of resistance in many bacteria. With a more specific diagnosis, a physician could prescribe a “narrow spectrum” antibiotic. Such an antibiotic would kill a limited number of bacterial species more precisely. This would slow the evolution of resistance.
Beyond the clinic, farmers use antibiotics to treat their animals and to prevent crop disease. This exposes microorganisms on the farm to antibiotics and could drive the evolution of resistance. Some livestock producers even use antibiotics to boost the growth of their animals whether they have infections or not. The WHO recommends putting a stop to this growth-promoting use. In the US, the FDA has rules intended to prevent the growth-promoting use of antibiotics .
If a region doesn’t have adequate systems for the disposal of human and animal waste, this can also encourage the spread of antibiotic resistance. When overwhelmed by waste streams, these inadequate systems may allow antibiotic resistant microorganisms to spread into water supplies. For example, sewage might leak into a river commonly used for bathing. Those bathing in the river might get antibiotic resistant infections as a result.
What are the mechanisms behind antibiotic resistance?
Microorganisms use many strategies to defend themselves against antibiotics. They can acquire genes encoding “efflux pumps” that eject antibiotics from their cells. They can evolve mutated versions of the proteins that the antibiotics normally interact with. They can even obtain genes encoding proteins that destroy antibiotics.
Regardless of the mechanism, the genes encoding antibiotic resistance are easily shared between microorganisms. For example, these genes may be found on extrachromosomal pieces of DNA called plasmids. Plasmids spread through microbial mating and can even be taken up from the environment. Viruses can also spread resistance genes between microorganisms.
CRISPR diagnostics and antibiotic stewardship
As we discussed in “CRISPR diagnostics in the fight against infectious disease,” one of the primary ways CRISPR diagnostics can improve antibiotic stewardship is by enabling physicians to prescribe the right drugs for the right infections. By identifying key nucleic acids, CRISPR diagnostics can pinpoint the organismal causes of infections. For example, CRISPR diagnostics might tell physicians whether an infection is caused by a virus or a particular bacterium. If the infection is viral, the physician won’t prescribe an antibiotic. This could slow the evolution of resistance.
CRISPR diagnostics can also tell physicians which antibiotics a microorganism is resistant to. For example, one could program a CRISPR diagnostic to detect an efflux pump. If detected, physicians would know to avoid prescribing antibiotics ejected by that pump.
Finally, CRISPR diagnostics could be used to monitor the environment for antibiotic resistant microorganisms. For example, public health researchers might take samples from a river commonly used for bathing. By testing these samples with a CRISPR diagnostic, researchers could determine whether or not the river harbors antibiotic resistant bacteria. Precautions could then be taken to avoid prescribing ineffective antibiotics to anyone in contact with the river. Officials could also prioritize river clean-up if a particularly dangerous microorganism were detected by the CRISPR diagnostic.
With CRISPR’s help, we can effectively fight antibiotic resistance!
Interested in joining the Mammoth team? Check out our open positions!
